Segmentation Model-Part IV - Data augmentation on the GPU with Kornia library
The fourth part of the Segmentation Tutorial Series, a step-by-step guide to developing data augmentation on GPU with Kornia library
- 1. Problem Description and Dataset
- 2. Data Preparation
- 3. The CPU bottleneck
- 4. Data Augmentation using Kornia
In this post, we discover how to use Kornia modules in order to perform the data augmentation on the GPU in batch mode. Kornia is a differentiable library that allows classical computer vision to be integrated into deep learning models. Kornia consists a lot of components. One of them is kornia.augmentation - a module to perform data augmentation in the GPU.
We will work with the Segmentation Problem (Nail Segmentation). For that, we use Pytorch Lightninig to train model and use Kornia to build the data augmentation on the GPU.
1. Problem Description and Dataset
We want to cover a nail semantic segmentation problem. For each image, we want to detect the segmentation of the nail in the image.
| Images | Masks |
|---|---|
 |
 |
Our data is organized as
├── Images
│ ├── 1
│ ├── first_image.png
│ ├── second_image.png
│ ├── third_image.png
│ ├── 2
│ ├── 3
│ ├── 4
├── Masks
│ ├── 1
│ ├── first_image.png
│ ├── second_image.png
│ ├── third_image.png
│ ├── 2
│ ├── 3
│ ├── 4
We have two folders: Images and Masks. Images is the data folder, and Masks is the label folder, which is the segmentations of input images. Each folder has four sub-folder: 1, 2, 3, and 4, corresponding to four distribution patterns of nail .
We download data from link and put it in data_root, for example
data_root = "./nail-segmentation-dataset"
2. Data Preparation
Similar to the training pipeline of the previous post, we first make the data frame to store images and masks infos.
| index | images |
|---|---|
| 1 | path_first_image.png |
| 2 | path_second_image.png |
| 3 | path_third_image.png |
| 4 | path_fourth_image.png |
For that we use make_csv_file function in data_processing.py file.
3. The CPU bottleneck
The fact is that today these transforms are applied one input at a time on CPUs. This means that they are super slow.
3.1 A naive approach model training

The naive training pipeline includes:
- The pre-processing of the data occurs on the CPU
- The model will be typically trained on GPU/TPU.
3.2 Data Augmentation using GPU
To improve the training speed we can shift the data augmentation task in to GPU
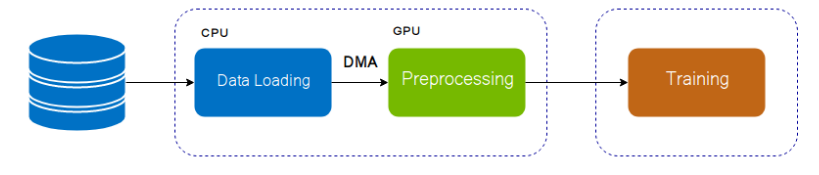
To do that we can use Kornia.augmentation, Dali libraries.
- Kornia.augmentation is the module of Kornia which permit to do augmentation in GPU. It will boost the speed of traininig in almost cases.
- DALI is the library for data loading and pre-processing to accelerate deep learning applications. Data processing pipelines implemented using DALI can easily be retargeted to TensorFlow, PyTorch, MXNet and PaddlePaddle.
This post we will focus on how to use Kornia. A guide of using DALI will be introduced in next post.
4. Data Augmentation using Kornia
In this part, we will cover how to use Kornia for data augmentation. To augumentate data on GPU, we use transforms (augumentations) as a transform_module ( in Pytorch platform, it is a nn.Module object) whose input is a tensor of size $C\times H \times W$ and output is also tensor of size $C\times H \times W$.
That transform_module is put between the processing task (includes read images, make images of batch having same size, convert images in to the tensor format) and the training model. More precisely,
class ModelWithAugumentation(nn.Module):
"""Module to perform data augmentation on torch tensors."""
def __init__(self, transform_module: nn.Module, model : nn.Module) -> None:
super().__init__()
self.transform_module = transform_module
self.model = model
def forward(self, x: Tensor) -> Tensor:
augmented_x = self.transform_module(x) # BxCxHxW
x_out = self.model(augmented_x)
return x_out
where transform_module is defined by using Kornia or torchvision. For example
transform_module = K.augmentation.AugmentationSequential(
K.augmentation.Normalize(Tensor((0.485, 0.456, 0.406)), Tensor((0.229, 0.224, 0.225)), p=1)
)
Note: we can also use torchvsiion to define the transform module.
transforms = torch.nn.Sequential(
transforms.CenterCrop(10),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225)),
)
Each transform module is a nn.Module object. We can use nn.Sequential to define a sequence of transforms.
We now apply that strategy to our problem. Comparing with the previous pipeline in the last post (Training deep learning segmentation models in Pytorch Lightning), here we have some modifications:
- Only use Resize or Padding in the data augmentation on CPUs, in the last part we define the whole augmentation by using albumentations and use it as the transform before going to the model.
import albumentations as A
def resize(p: float = 1):
return A.Resize(384, 384, always_apply=True)
self.valid_transform = resize()
self.train_transform = resize()
- Using Kornia to define the augmentation (nn.Module object), here we have
train_transform_Kandvalid_transform_K
import kornia as K
valid_transform_K = K.augmentation.AugmentationSequential(
K.augmentation.Normalize(Tensor((0.485, 0.456, 0.406)), Tensor((0.229, 0.224, 0.225)), p=1),
data_keys=["input", "mask"],
)
train_transform_K = K.augmentation.AugmentationSequential(
K.augmentation.container.ImageSequential( # OneOf
K.augmentation.RandomHorizontalFlip(p=0.6),
K.augmentation.RandomVerticalFlip(p=0.6),
random_apply=1,
random_apply_weights=[0.5, 0.5],
),
K.augmentation.ColorJitter(0.1, 0.1, 0.1, 0.1, p=0.5),
# K.augmentation.RandomAffine( degrees = (-15.0,15.0), p= 0.3),
K.augmentation.Normalize(Tensor((0.485, 0.456, 0.406)), Tensor((0.229, 0.224, 0.225)), p=1),
data_keys=["input", "mask"],
same_on_batch=False,
)
- In the LightningModule, we define two new functions (or two nn.Module objects)
self.train_transform = train_transform_K
self.valid_transform = valid_transform_K
Note: Add transform into the training loop and the valid loop (
training_stepandvalidation_step)
def training_step(self, batch, batch_idx):
imgs, masks = batch["image"], batch["label"]
if self.train_transform is not None:
imgs, masks = self.train_transform(imgs, masks) # add the transform before going to the model
imgs, masks = imgs.float(), masks.float()
logits = self(imgs)
train_loss = self.loss_function(logits, masks)
train_dice_soft = self.dice_soft(logits, masks)
self.log("train_loss", train_loss, prog_bar=True)
self.log("train_dice_soft", train_dice_soft, prog_bar=True)
return {"loss": train_loss, "train_dice_soft": train_dice_soft}
def validation_step(self, batch, batch_idx):
imgs, masks = batch["image"], batch["label"]
if self.valid_transform:
imgs, masks = self.valid_transform(imgs, masks) # add the transform before going to the model
imgs, masks = imgs.float(), masks.float()
logits = self(imgs)
valid_loss = self.loss_function(logits, masks)
valid_dice_soft = self.dice_soft(logits, masks)
valid_iou = binary_mean_iou(logits, masks)
self.log("valid_loss", valid_loss, prog_bar=True)
self.log("valid_dice", valid_dice_soft, prog_bar=True)
self.log("valid_iou", valid_iou, prog_bar=True)
return {
"valid_loss": valid_loss,
"valid_dice": valid_dice_soft,
"valid_iou": valid_iou,
}
We keep all of rest parts of the pipeline (LightningDataModule, Trainer).
For more details, we can find the source code at github